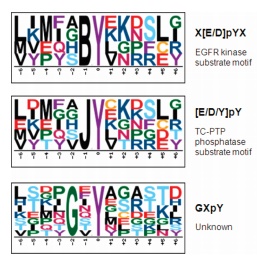
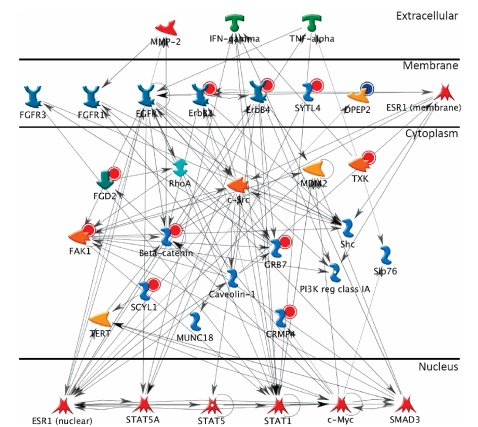
Today, Xiaobian talks to you about phosphorylated proteome identification ! Protein post-translational modifications (PTMs) are involved in almost all of the normal life activities of cells and play a very important regulatory role. Protein modification has become an extremely important field in protein research in the world. Currently, there are more mature phosphorylation, acetylation, glycosylation and ubiquitination. Protein phosphorylation is the most common and important post-translational modification of proteins in organisms. It can participate in the regulation of growth, development, stress, disease and other life by stimulating and regulating many signaling pathways. The process has always been the focus and hotspot of biological research. According to customer needs, Jin Kairui proteome platform can provide full-spectrum identification of phosphorylated proteome and label-free quantitative technical services. The full-spectrum identification of phosphorylated proteomes is based on more complex samples such as tissues and cells, and aims to identify phosphorylated proteins and corresponding phosphorylation sites in the samples. First, the protein sample was enzymatically digested, TiO2 or IMAC-Fe or IMAC-Ti was enriched for phosphorylated polypeptide, and 5600-plus mass spectrometry was used to compare the obtained mass spectrogram with the corresponding database search to obtain the peptide sequence result. The bioinformatics software calculates the phosphorylation site. For the identification of phosphorylation sites, in order to increase the accuracy of localization of the modified sites, Jin Kairui uses the more popular Ascore algorithm to further score the phosphorylation modifications occurring at each point to correctly identify the true modification sites. Technical route: Technical features: â— The enrichment method has high specificity, is highly resistant to low pH solutions, detergents, salts, and other low molecular contaminants, and is easily separated from non-phosphorylated peptides; â— Large flux, more than 1000 phosphorylation sites can be identified at one time. Scope of application: â— Known species genome sequence, ESTs sequence or protein sequence library; â— No other special requirements. Classic Case: Title: Identification of tyrosine-phosphorylated proteins associated with lung cancer metastasis using label-free quantitative analyses. Identification of tyrosine phosphorylation proteins associated with lung cancer metastasis using Label-free quantitative techniques Journal: journal of proteome research Main technology: Label-free quantitative technology Abstract: Tyrosine phosphorylation (P-tyrosine) protein may be involved in the invasion and metastasis of lung cancer, but it has been reported to be less. In this paper, the tyrosine phosphorylation protein and its involvement in the metastasis process were studied by Label-free quantitative technique. A total of 335 P-Tyr sites from 276 phosphorylated proteins were identified. Label-free quantitative results showed that 36 phosphorylated peptides showed significant differences between samples with different tyrosine phosphorylation protein invasive ability. Two new site conserved sequences and four known specific kinase and phosphorylated motif sequences were extracted from these peptides: EGFR, Src, JAK2 and TC-PTP. Analysis of protein interactions with P-tyrosine proteins revealed that 11 proteins were linked in an interaction network containing EGFR, c-Src, c-Myc and STAT, which has been linked to lung cancer metastasis. Seven of these 11 proteins have not been previously reported, which will provide a better and comprehensive understanding of the mechanisms and treatments for lung cancer invasion/metastasis. Figure: Three newly shown motifs Wu HY, Tseng VS et al. Identification of tyrosine-phosphorylated proteins associated with lung cancer metastasis using label-free quantitative analyses. Journal of proteome research Figure: Network interaction map of tyrosine phosphorylated proteins with significant changes in expression Wu HY, Tseng VS et al. Identification of tyrosine-phosphorylated proteins associated with lung cancer metastasis using label-free quantitative analyses. Journal of proteome research References: Wu HY, Tseng VS, Chen LC, Chang HY, Chuang IC, Tsay YG, Liao PC: Identification of tyrosine-phosphorylated proteins associated with lung cancer metastasis using label-free quantitative analyses. Journal of proteome research 2010, 9( 8): 4102-4112. Insulin Syringes Needle,Disable Syringe,Monoject Syringe,10 Ml Syringe FOSHAN PHARMA CO., LTD. , https://www.fs-pharma.com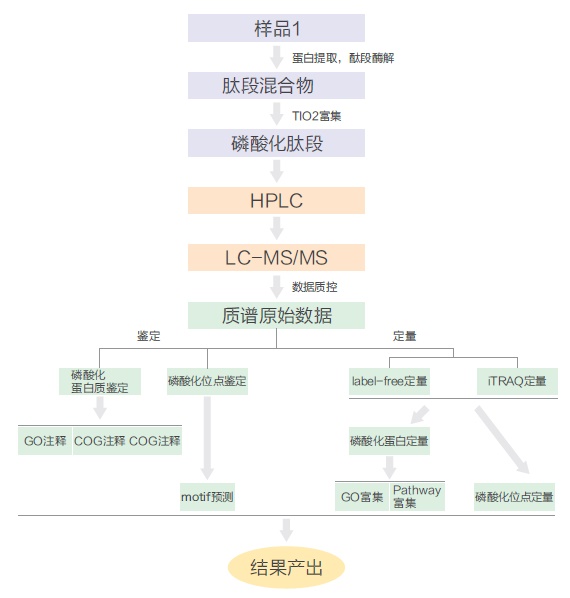
Phosphorylated proteome identification technology route and classic case introduction